- Main concepts
This research is inspired by the fact that artificial neural networks (NN) have proved their efficiency in pattern recognition, signal processing and ECG interpretation. Following that fact, the authors develop a new original approach for QRS segmentation based on 2-stage 1-dimensional convolutional neural network and perform the comparison of this algorithm with several other deep learning models, since it is impossible to make a monosemantic choice in favor of a particular NN architecture without thorough comparative study. As a result, the obtained algorithm is deployed into HM software using specially designed conversion method, which utilizes CPU-built extensions for effective performance of the algorithm.
The development of QRS segmentation system goes through the steps represented in Figure 1. The research is aimed to define the most precise signal segmentation algorithm with the least computing power consumption.
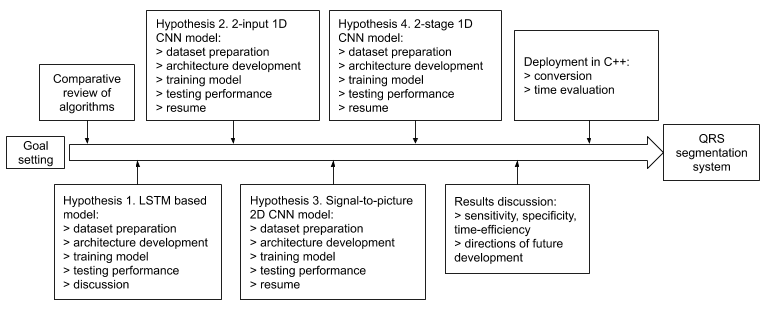
Figure 1. R&D pipeline
For an objective comparison of the algorithms’ effectiveness, it is necessary to evaluate their work in the same conditions using the same criteria. In this regard, the authors have tested four algorithms (hereinafter hypotheses) in the same way using the same datasets. As part of testing the first hypothesis, LSTM-based model with pre- and post-processing operations is developed and dataset for training is formed. After training the model, testing takes place. According to the acquired results, the hypothesis about the applicability of the approach is accepted or rejected. At the next stage, the same sequence of operations is applied to the 2nd hypothesis about 2-input 1-dimensional convolutional neural network model. Hypothesis 3 about “signal-to-picture” approach based on 2-dimensional convolutional model is checked. Finally, the drawbacks of previous approach are considered, and the fourth method based on 2-stage 1-D CNN approach is developed. After aggregating the performance characteristics of each method, the best approach is defined, deployment in C++ HM ECG analysis software is carried out, and directions of model development are discussed.
- Research outcomes
For further discussion three time-efficient approaches with the highest accuracy are selected, leaving aside signal segmentation pipeline based on the BLSTM model. According to the models’ evaluation results, dramatic increase in time consumption is observed in case of “signal-to-picture” 2-D CNN approach. However, this method requires bulky data preprocessing step that significantly loads the system. To get around this computationally intensive stage, 2-stage 1-D CNN approach is developed. The main intuitions of the hypothesis 3 are implemented in hypothesis 4 taking into account the transition from two-dimensional to one-dimensional architecture. Along with much simpler data preprocessing this approach requires only 0.01 second for analysis of 1 second of 360 Hz ECG record. Moreover, it gives sufficient accuracy which is comparable to other approaches. The evaluation results are shown in Table 1.
Table 1. Comparison of three most suitable approaches

2-stage 1-D CNN approach utilizes the simplest possible one-dimensional convolutional neural networks in two steps: at first for peak location, and then for edge detection. It allows to reduce computing power requirements, hence increase time-efficiency. Detailed description of the model evaluation on MIT-BIH DB for peak and edge detection is represented in the original research paper [LINK to the FRUCT publication].
- Deployment
The aim of this step is to integrate the developed ECG segmentation model into the HM ECG analysis software. The main problem is that usually there is no personal computer with either integrated or discrete graphical processing unit (GPU) at the typical medical institution. In this regard, it is necessary to implement an approach that allows to perform all the operations only with central processing unit (CPU). 2-stage 1-D CNN approach (hypothesis 4) is chosen as the most efficient method for implementation in C++.
The authors have applied several different CPU-built extensions in order to provide time-efficiency of the algorithm. Universal approach consisting in automatic selection of the fastest extension supported by the particular CPU have been developed. Comparison of processing time using different CPU-built extensions is shown below in Table 2.
Table 2. Comparison of processing time using different CPU-built extensions
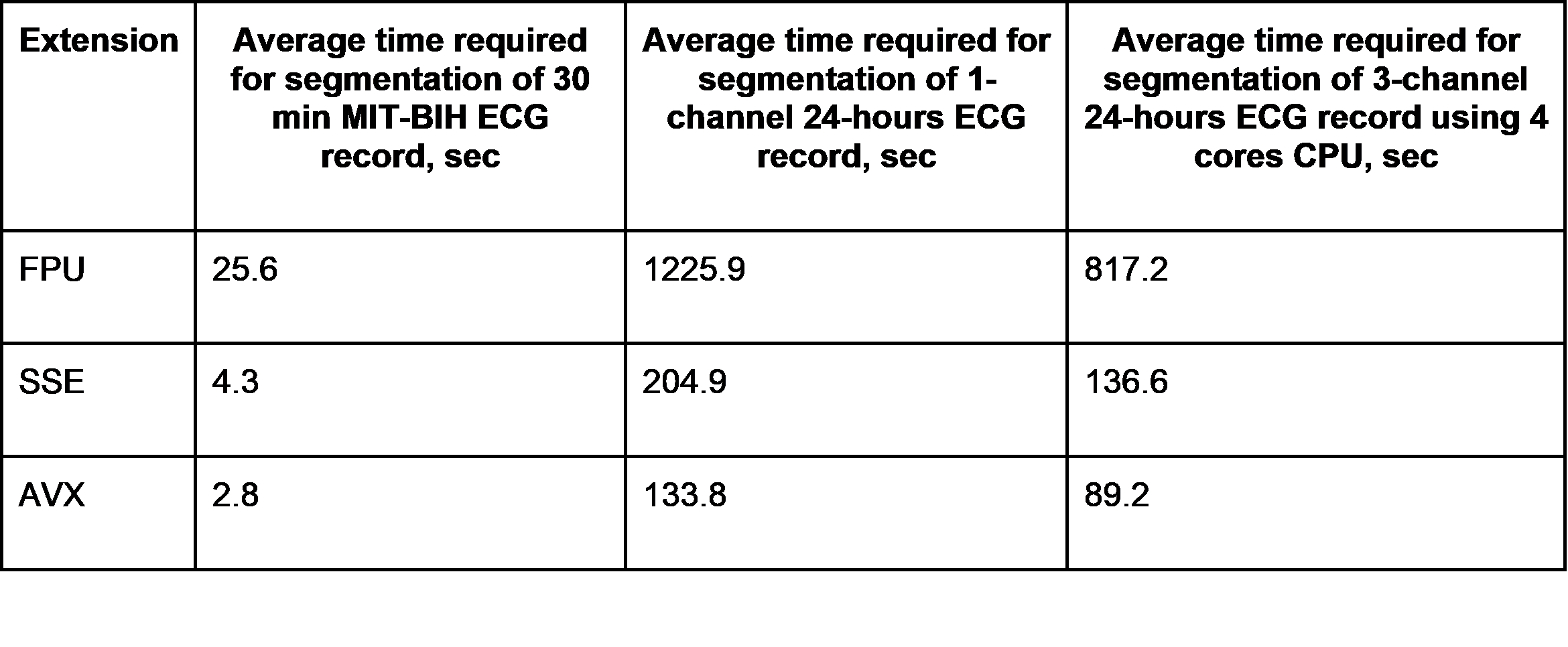
- Summary
In the course of the research four hypotheses about ECG segmentation methods are checked and the best one is accepted. BLSTM-based model is built and tested, but evaluation shows the impossibility of applying the model in real-life applications. 2-input 1-D CNN model has demonstrated high accuracy of peak and edges detection – average accuracy is 99.18%, but long processing time – it takes 0.45 seconds to process 1 second of 360 Hz MIT DB ECG record. “Signal-to-picture” approach has proved its efficiency in terms of accuracy – 99% which is close to the results of the previous approach, and time-efficiency – the proposed algorithm is 15 times faster than the previous one. Although it seems to be the most appropriate approach for QRS segmentation, it has one major disadvantage which is data preprocessing step. ECG signal registered in time domain form needs to be transformed into 2-D data, in turn it leads to time and computing power consumption at the data preparation step. Finally, 2-stage 1-D CNN model has shown a bit lower accuracy characteristic, although significantly less time and computing power consumption. This method is chosen for deployment into HM ECG analysis software. The following technical solutions are applied for better performance of the algorithm operation on CPU: CPU-built extensions including FPU, SSE, AVX. As a result, the best averaged processing time of 3-channel 24-hours ECG record performed by using AVX extension is 1.5 minutes.
For more detailed information please refer to the original research paper.
